The Switzer lab uses biochemical and biophysical tools to investigate enzyme function. We study enzymes that a play a role in human disease. Our current work examines enzymes involved in pyrimidine metabolism. We are studying two enzymes: DNA methyltransferases and dihydroorotate dehydrogenases. There is more info below about each project.


 Students in the lab get hands-on experience with common molecular biology, biochemical, and biophysical techniques. Student projects often include techniques such as site-directed mutagenesis and/or molecular cloning, protein expression and purification, fluorescence anisotropy, various kinetics assays, ligand binding assays, circular dichroism spectroscopy, and isothermal titration calorimetry. If you are interested in pursuing research in the lab, please contact Professor Switzer.
Students in the lab get hands-on experience with common molecular biology, biochemical, and biophysical techniques. Student projects often include techniques such as site-directed mutagenesis and/or molecular cloning, protein expression and purification, fluorescence anisotropy, various kinetics assays, ligand binding assays, circular dichroism spectroscopy, and isothermal titration calorimetry. If you are interested in pursuing research in the lab, please contact Professor Switzer.
DNA Methyltransferases
DNA functions to store genetic information, which provides the sequence for protein synthesis. All of your cells contain the same genetic information, i.e. DNA sequence. Yet, we are made up of a multitude of specialized cells. This diversity is accomplished by activating expression of some genes while inhibiting expression of others. Epigenetics is the study of heritable changes in genome function that do not alter the underlying DNA sequence. Common epigenetic mechanisms that regulate gene expression include DNA methylation and post-translational modification of histone proteins. Thus, while the genetic information provides the sequence for proteins, the epigenetic information provides instructions on when and where the proteins will be expressed.
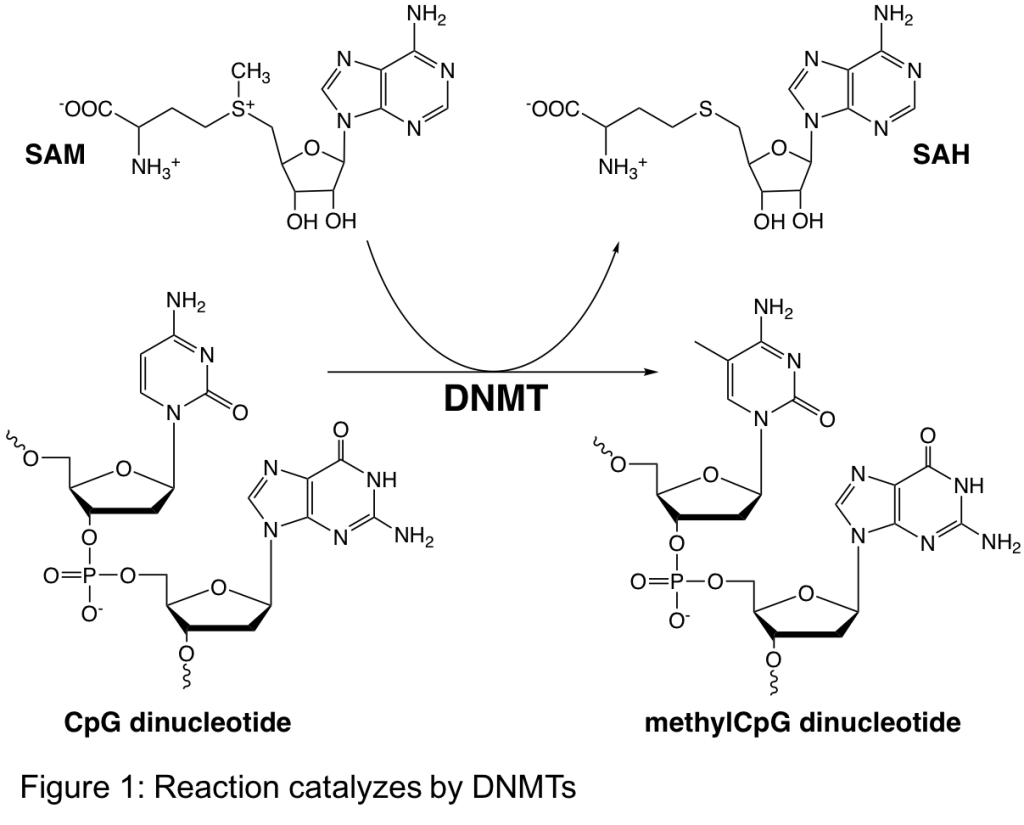
In humans, the most common epigenetic DNA modification is methylation of the 5-carbon of cytosines, predominately in CpG dinucleotides (Figure 1). Cytosine methylation is a repressive epigenetic mark that results in gene silencing. Methylation patterns are established and maintained by a family of enzymes known as DNA methyltransferases (DNMTs). Changes to the normal methylation pattern have been associated with several diseases including neurodegenerative diseases and cancer. Understanding the activity, regulation, and inhibition of these enzymes in vitro will aid in our understanding of their biological function in vivo as well as their role in various diseases.
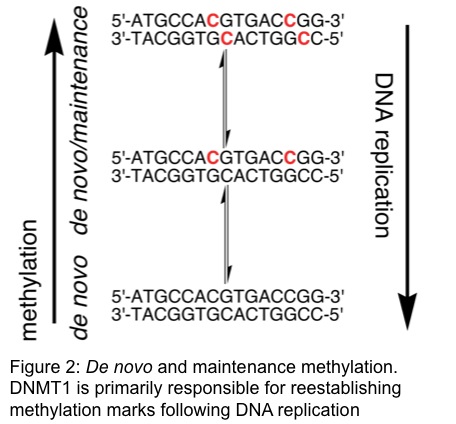
There are three catalytically active isoforms of DNMTs in humans – DNMT3a, DNMT3b, and DNMT1. Methylation patterns are established by DNMT3a and DNMT3b. Since DNA replication is a semi-conservative process, one strand of the duplex DNA is devoid of methylation marks following cell division. Methylation patterns are primarily maintained by DNMT1, which is the most abundant DNMT and possesses specificity for methylation of this hemimethylated DNA (Figure 2). We are currently investigating the biochemical consequence of disease-associated mutations in DNMT1. We are also currently working to identify and characterize novel DNMT1 inhibitors.
Dihydroorotate Dehydrogenases
Dihydroorotate dehydrogenases (DHODs) are flavin-containing enzymes that catalyze the fourth step in de novo pyrimidine biosynthesis. Pyrimidines are important biomolecules involved in DNA replication, transcription, lipid and complex sugar biosynthesis, and more. Inhibiting pyrimidine biosynthesis can lead to cell death, making DHOD a drug target.
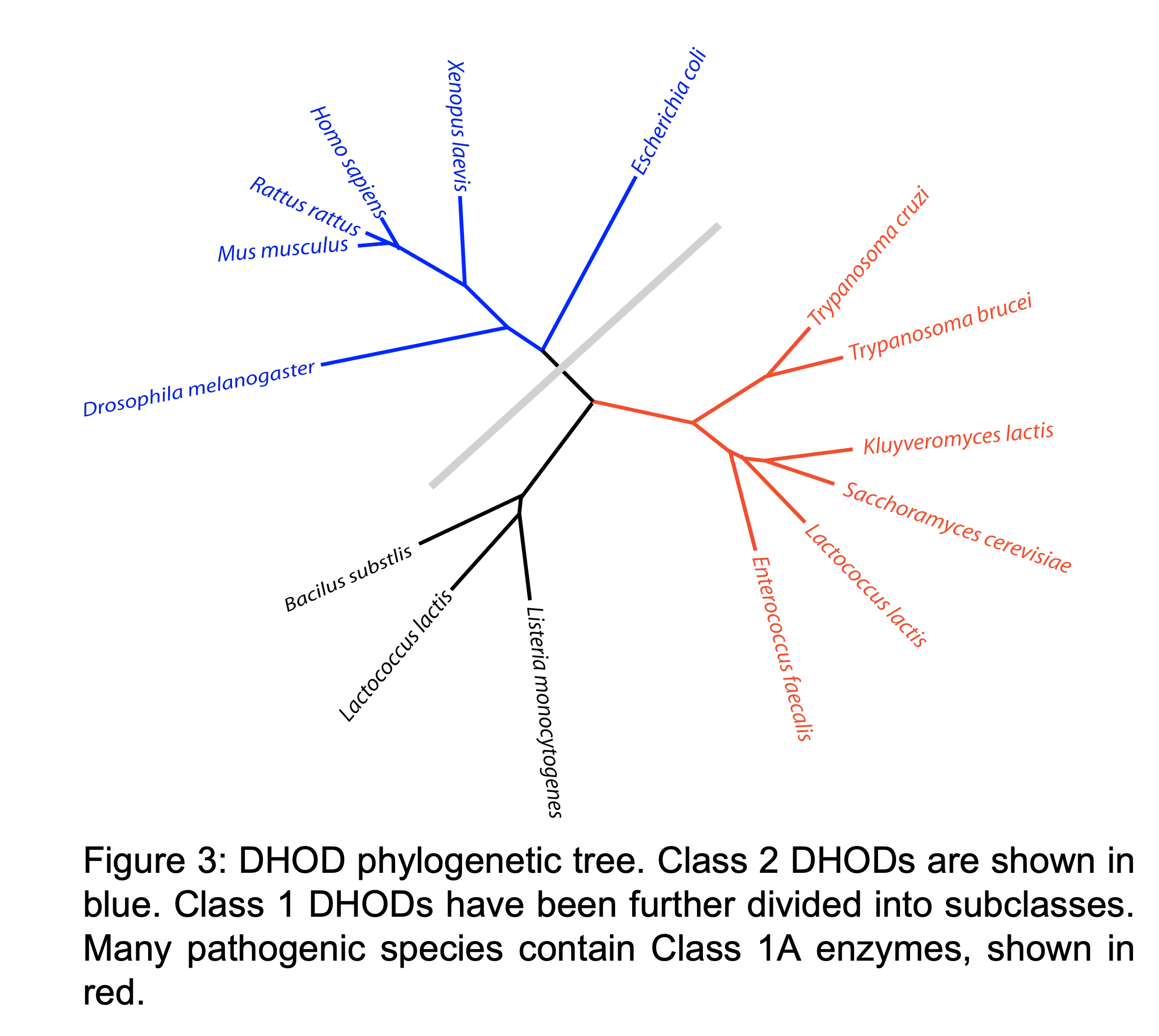 DHODs have been divided into classes based on sequence. Humans contain a Class 2 DHOD, while several pathogenic species including E. faecalis and T. cruzi contain Class 1A DHODs (Figure 3). Therefore, inhibitors that bind specifically to Class 1A DHODs could be used to treat a variety of diseases with little to no side effects in humans. Even though the pyrimidine binding site in all DHODs is nearly identical, small molecules that bind only to Class 1A DHODs have been discovered. The underlying reason(s) for this specificity remain a mystery. We are using site-directed and random mutagenesis to investigate the structural features critical for inhibitor binding to Class 1A DHODs. Steady-state kinetics, ligand binding assays, and CD spectroscopy are being used to characterize mutant enzymes.
DHODs have been divided into classes based on sequence. Humans contain a Class 2 DHOD, while several pathogenic species including E. faecalis and T. cruzi contain Class 1A DHODs (Figure 3). Therefore, inhibitors that bind specifically to Class 1A DHODs could be used to treat a variety of diseases with little to no side effects in humans. Even though the pyrimidine binding site in all DHODs is nearly identical, small molecules that bind only to Class 1A DHODs have been discovered. The underlying reason(s) for this specificity remain a mystery. We are using site-directed and random mutagenesis to investigate the structural features critical for inhibitor binding to Class 1A DHODs. Steady-state kinetics, ligand binding assays, and CD spectroscopy are being used to characterize mutant enzymes.